Difference between revisions of "AutoMeKin"
(→Program execution) |
(→References and citations) |
||
| Line 88: | Line 88: | ||
<div style="column-count:2;-moz-column-count:2;-webkit-column-count:2"> | <div style="column-count:2;-moz-column-count:2;-webkit-column-count:2"> | ||
<ol start="5"> | <ol start="5"> | ||
| + | <li>[https://pubs.acs.org/doi/abs/10.1021/acs.jpca.1c00690 <span style="font-size:100%"> Ferreira da Silva et al. J. Phys. Chem. A 2021, (https://doi.org/10.1021/acs.jpca.1c00690)</span>]</li> | ||
<li>[https://www.sciencedirect.com/science/article/pii/S0040402019311573 <span style="font-size:100%"> A. Esteban et al. Tetrahedron 2020, 76, 130764</span>]</li> | <li>[https://www.sciencedirect.com/science/article/pii/S0040402019311573 <span style="font-size:100%"> A. Esteban et al. Tetrahedron 2020, 76, 130764</span>]</li> | ||
<li>[https://onlinelibrary.wiley.com/doi/full/10.1002/syst.201900024 <span style="font-size:100%"> R. A. Jara-Toro et al. ChemSystemsChem 2020, 2, e1900024</span>]</li> | <li>[https://onlinelibrary.wiley.com/doi/full/10.1002/syst.201900024 <span style="font-size:100%"> R. A. Jara-Toro et al. ChemSystemsChem 2020, 2, e1900024</span>]</li> | ||
Revision as of 12:55, 16 March 2021
Contents
Introduction
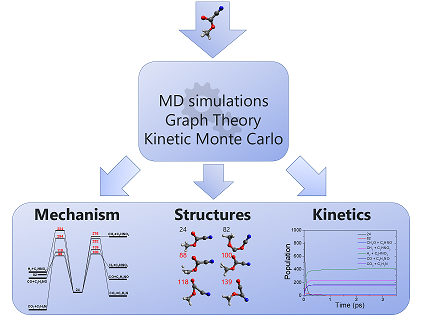
AutoMeKin (formerly tsscds) has been designed to discover reaction mechanisms in an automated fashion. Transition states are located using MD simulations and Graph Theory algorithms. Monte Carlo simulations afford kinetic results. The only input is a starting structure in XYZ format. The method is described in these two publications: 1 2. At present MOPAC2016, Entos Qcore and Gaussian 09 (G09) are interfaced with AutoMeKin. The program has been tested on the following Linux distros: CentOS 7, Red Hat Enterprise Linux and Ubuntu 16.04.3 LTS.
To give you a flavor of the capabilities of the program you can try our web interface
Authors
George L. Barnes, David R. Glowacki, Sabine Kopec, Emilio Martinez-Nunez, Daniel Pelaez-Ruiz, Aurelio Rodriguez, Roberto Rodriguez-Fernandez, Robin J. Shannon, James J. P. Stewart, Pablo G. Tahoces and Saulo A. Vazquez
Address
Departamento de Química Física, Facultade de Química, Avda. das Ciencias s/n, 15782 Santiago de Compostela, SPAIN
email me
License
MIT License
Copyright (C) 2021 George L. Barnes, David R. Glowacki, Sabine Kopec, Emilio Martinez-Nunez, Daniel Pelaez-Ruiz, Aurelio Rodriguez, Roberto Rodriguez-Fernandez, Robin J. Shannon, James J. P. Stewart, Pablo G. Tahoces and Saulo A. Vazquez
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
Install the code
To install the code follow the Installation instructions
Tutorial
Download tutorial
Program execution
Unless you donwloaded the singularity container (in that case skip this step), to start using any of the scripts of the program, load the amk/2021 module:
module load amk/2021
To run the low-level calculations use:
nohup llcalcs.sh molecule.dat ntasks niter runningtasks >llcalcs.log 2>&1 &
where:
molecule is the name of your molecule
ntasks is the number of tasks
niter is the number of iterations
runningtasks is the number of simultaneous tasks
To run the high-level calculations use:
nohup hlcalcs.sh molecule.dat runningtasks >hlcalcs.log 2>&1 &
For more details, follow the instructions given in the tutorial
References and citations
These four publications must be cited in any work presenting results obtained with our software:
- E. Martínez-Núñez J. Comput. Chem. 2015, 36, 222
- E. Martínez-Núñez Phys. Chem. Chem. Phys. 2015,17, 14912
- A. Rodriguez, R. Rodriguez-Fernandez, S.A. Vazquez, G.L. Barnes, J.J.P. Stewart, E Martínez-Nuñez, J. Comput. Chem. 2018, 39, 1922
- J. J. P. Stewart, MOPAC2016, Stewart Computational Chemistry: Colorado Springs, CO, USA, HTTP://OpenMOPAC.net, 2016.
Publications using AutoMeKin:
- Ferreira da Silva et al. J. Phys. Chem. A 2021, (https://doi.org/10.1021/acs.jpca.1c00690)
- A. Esteban et al. Tetrahedron 2020, 76, 130764
- R. A. Jara-Toro et al. ChemSystemsChem 2020, 2, e1900024
- R. Panades-Barrueta et al. Frontiers in Chemistry 2019, 7, 576
- S. Kopec et al. Int. J. Quantum Chem. 2019, 119, e26008
- V. Macaluso et al. J. Phys. Chem. A 2019, 123, 3685-3696
- S. A. Vázquez et al. Molecules 2018, 23, 3156
- A. Rodríguez et al. J. Comput. Chem. 2018, 39, 1922-1930
- D. Ferro-Costas et al. J. Phys. Chem. A 2018, 122, 4790-4800
- Y. Fenard et al. Combust. Flame. 2018, 191, 252-269
- M. J. Wilhelm et al. ApJ. 2017, 849, 15
- J. A. Varela et al. Chem. Sci 2017, 8, 3843-3851
- E. Rossich-Molina et al. Phys. Chem. Chem. Phys. 2016, 18, 22712-22718
- R. Pérez-Soto et al. Phys. Chem. Chem. Phys. 2016, 18, 5019-5026
- S. A. Vázquez and E. Martínez-Núñez, Phys. Chem. Chem. Phys. 2015, 17, 6948-6955
Changelog
Consult the Latest changes
Web interface
AutoMeKin can be used through our web interface.
News
The improvements briefly described below are available in AutoMeKin2021.
AutoMeKin has been interfaced with Entos Qcore
AutoMeKin has been interfaced with BXDE to enhance its efficiency ( R. A. Jara-Toro et al. ChemSystemsChem doi: 10.1002/syst.201900024).
The method has also been recently generalized in a collaboration with Dani Pelaez and co-workers to study van der Waals structures ( S. Kopec et al. Int. J. Quantum Chem. 2019, 119, e26008 ) and also to generate sum-of-products PESs for quantum dynamics ( R. Panades-Barrueta et al. Frontiers in Chemistry 2019, 7, 576).
Return to Contents
Return to Main_Page